MetagenomeScope
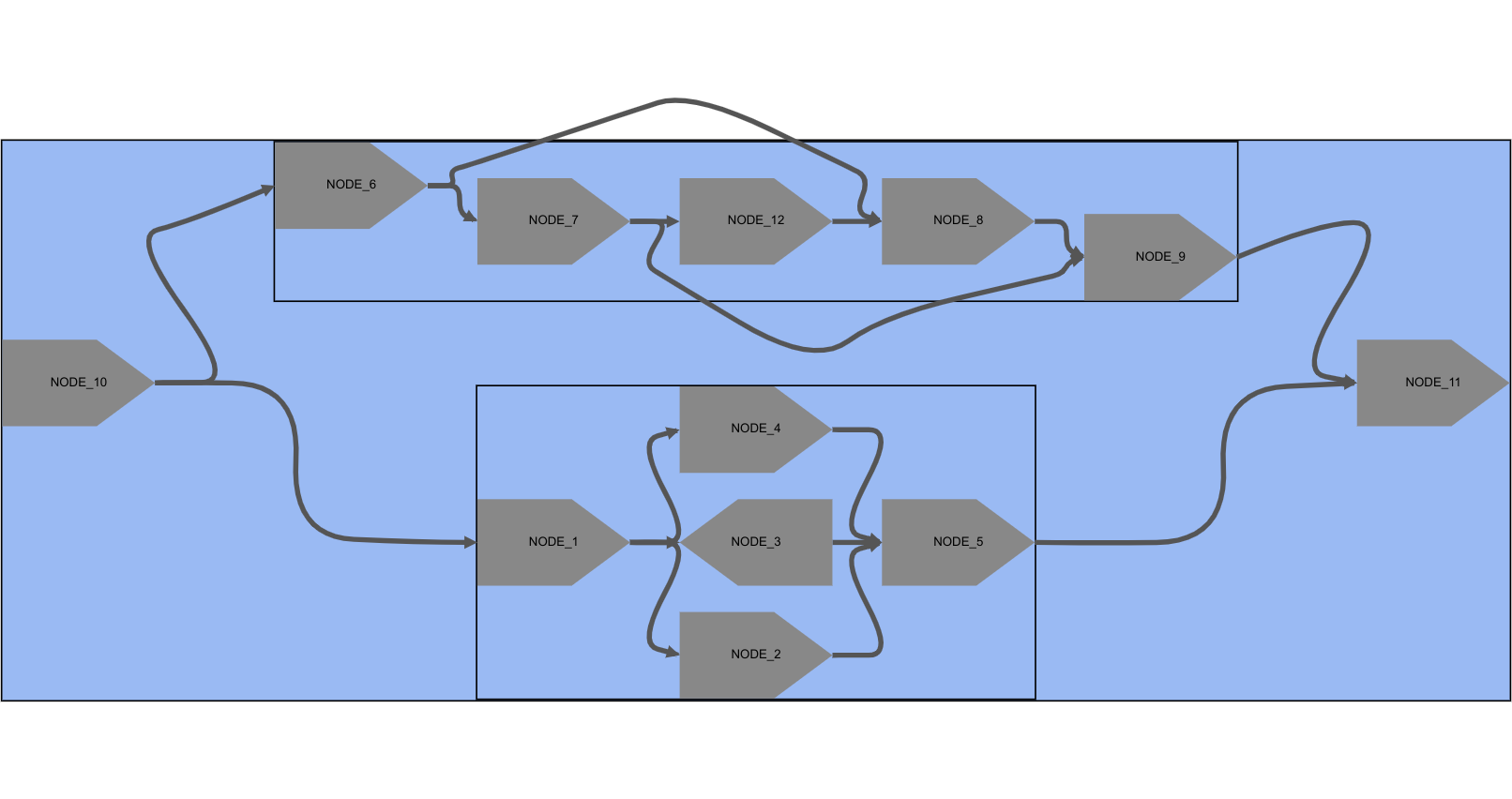
Visualization of a graph based on Fig. 2(a) of [Nijkamp et al. 2013].
Due to MetagenomeScope's use of hierarchical pattern decomposition, the input assembly graph is automatically annotated as a bubble containing two smaller bubbles: each of the three bubbles can be interactively collapsed and uncollapsed in the interface.
Check out a demo of this visualization!
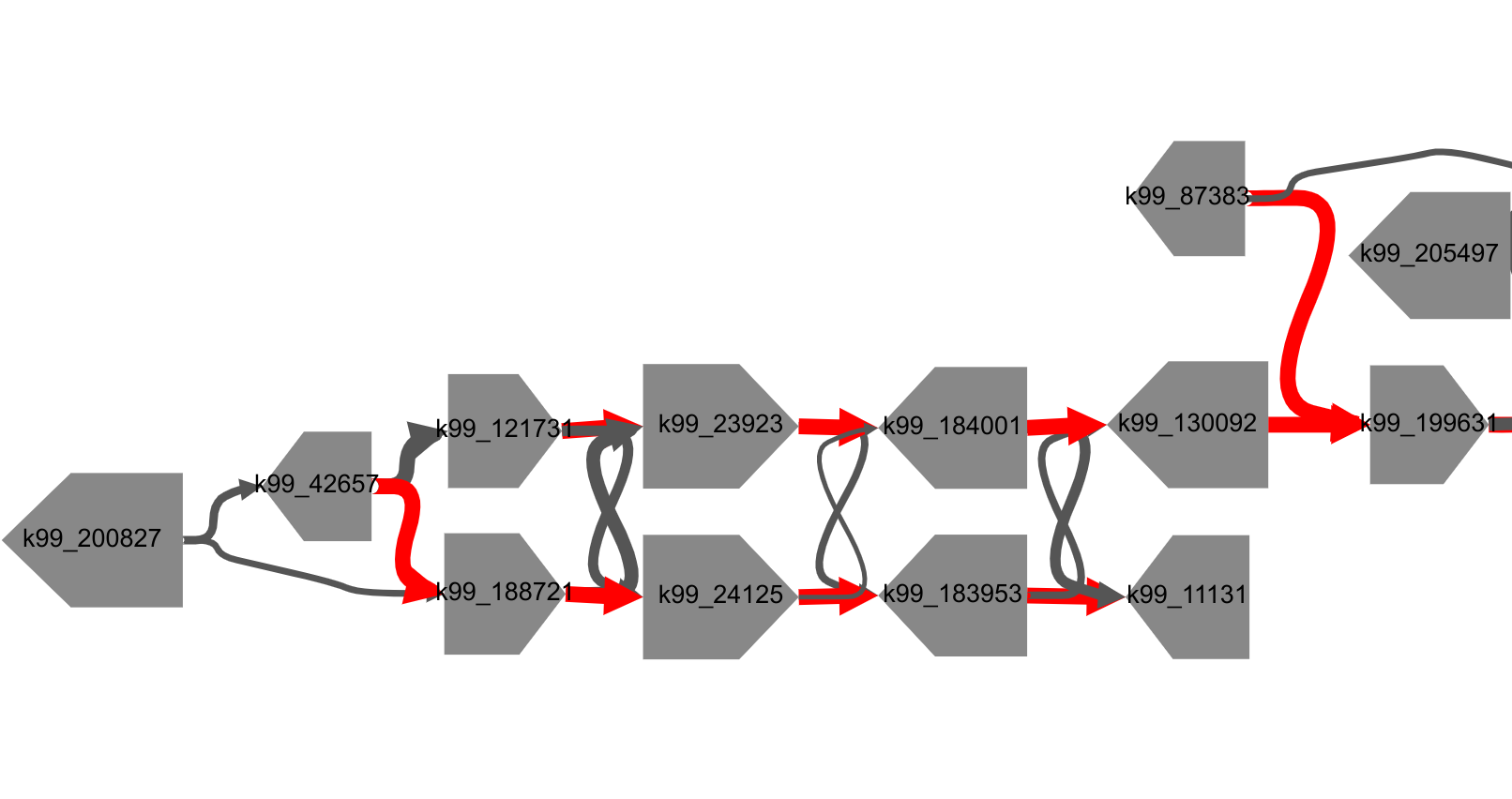
Visualization of a region of a stool metagenome scaffold graph produced by MetaCarvel [Ghurye et al. 2019].
MetagenomeScope performs hierarchical graph layout using Graphviz' dot layout tool. This simplifies the visualization and manual inspection of complex, small-scale details within assembly graphs, such as these branching paths.
News
December 2020: MetagenomeScope is back! We just merged in a new version of the codebase featuring many new features, including hierarchical pattern decomposition, bubble boundary duplication, drawing multiple components at once, and much more. Check out the code and installation instructions on GitHub!
August 2018: Gave a presentation and tutorial on MetagenomeScope during the 2018 Strategies and Techniques for Analyzing Microbial Population Structures (STAMPS) course at the Marine Biological Laboratory! Check out the slides here, and check out the command-line MetagenomeScope tutorial here.
January 2018: Presented a poster on MetagenomeScope at the 2018 Winter Mid-Atlantic Microbiome Meetup (M3)!
September 2017: Presented a poster on MetagenomeScope at GD 2017! Check out the most recent version of the poster here.
Overview
MetagenomeScope is a web-based visualization tool designed for metagenomic assembly graphs. MetagenomeScope aims to display a semi-linearized, hierarchical overview of the input graph while emphasizing the presence of certain structural patterns in the graph.
The tool consists of a preprocessing script (written in Python) and a visualization interface (a client-side web application). The preprocessing script takes as input an assembly graph file (currently LastGraph, GFA, FASTG, and MetaCarvel GML files are accepted) and creates a directory containing the HTML / JS / CSS visualization.
MetagenomeScope is still under active development, so please get in touch with us if you have any questions or other feedback about the tool.
Highlighted features
- Hierarchical graph layout performed using Graphviz' dot layout engine.
-
Hierarchical pattern decomposition:
identifies and supports interactive collapsing/uncollapsing
of bubbles and frayed ropes [Miller, Koren, Sutton 2010], along with linear and cyclic linear sequences of
contigs.
- Runs pattern decomposition iteratively, so that patterns containing other patterns (e.g. bubbles containing chains) can be identified. In the viewer interface, these patterns can be collapsed or uncollapsed to show the graph at different levels of "complexity."
-
Scales edges in the graph by their
multiplicity/bundle size [Miller, Koren, Sutton 2010].
- Uses Tukey fences to identify "outlier" edge weights and omit them from the scaling process
- Outlier edges are specially colored in the graph to indicate their unusual high/low weight
Code
The source code for MetagenomeScope is publicly available on GitHub here. MetagenomeScope is licensed under the GNU General Public License, version 3. (If you're interested, the source code for this site is also available on GitHub.)
See the acknowledgements section of MetagenomeScope's wiki for an up-to-date list of the third-party libraries used by MetagenomeScope.
Contact
MetagenomeScope was created by members of the Pop Lab in the Center for Bioinformatics and Computational Biology at the University of Maryland, College Park.
Feel free to email mfedarko (at) umd (dot) edu with
any questions, suggestions, comments, concerns, etc. regarding
the tool. If you'd like, you can also
open an issue
on MetagenomeScope's GitHub page.